Lindenmayer Systems in MGS
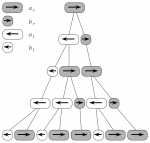
Lindenmayer Systems (or L-systems for short) are generative grammars working as a parallel sequence rewriting. A L-system corresponds to a sequence  of symbols transformed using rules of the form
of symbols transformed using rules of the form  so that if the sub-sequence
so that if the sub-sequence  is found in
is found in  , it is replaced by
, it is replaced by  . The rewriting strategy is maximal parallel meaning that rules all together through the sequence disjointly and maximally (i.e., such that there is no untouched subsequence containing a sub-sequence
. The rewriting strategy is maximal parallel meaning that rules all together through the sequence disjointly and maximally (i.e., such that there is no untouched subsequence containing a sub-sequence  ). L-systems are widely used for modeling biological systems and particularly for describing growth processes of plant development.
). L-systems are widely used for modeling biological systems and particularly for describing growth processes of plant development.
L-systems are by nature easily specified in MGS. In the following, we present two model of growth of Anabaena Catenula based on L-systems and implemented in MGS.
Anabaena Catenula
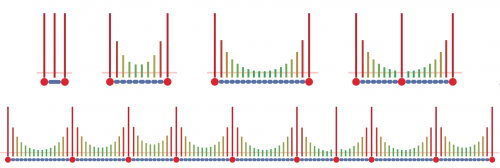
The cyanobacterium Anabaena grows in filaments of 100 cells or more. The growth process exhibits two interesting behaviors:
- Cell polarities: cells in the filament divide in a asymmetric manner: one of the daughters is longer than the other; more over the two possibilities for ordering the daughters (small-long or long-small) obey to a specific rule.
- Cell differentiation: when starved for nitrogen, specialized cells called heterocysts differentiate from the photosynthetic vegetative cells at regular intervals along each filament. Heterocysts are anaerobic factories for nitrogen fixation; in them, the nitrogenase enzyme complex is synthesized and the components of the oxygen-evolving photosystem are turned off. Plant signals exert both positive and negative regulatory control on heterocyst differentiation.
Simple Growth Model
Cells in the filament divide in a asymmetric manner: one of the daughters is longer than the other; more over the two possibilities for ordering the daughters (small-long or long-small) obey to a specific rule. A symbolic model of this behavior is described in The Algorithmic Beauty of Plants (pp. 5).
Cell state is symbolically abstracted with two properties:
- cell size: long or short
- cell polarity: left or right
Long cells divide quickly than short, and the polarity decide the division ordering (small-long or long-small).
MGS Implementation
Cells have polarities (Left or Right) and two different sizes (Short or Long)
type cell = `Left_Long | `Right_Long | `Left_Short | `Right_Short ;;
The whole alga is represented by a sequence of cells
type anabaena = [cell]seq ;;
Let define the initial state S0 as a sequence with a unique long right oriented cell
S0 := seq:(`Right_Long) ;;
Let check that the initial state is of type anabaena (dynamical typing)
anabaena(S0) ;;
Let define the evolution rules of the cells
trans grammar = { // Growth Rules: short cells become long while keeping their polarity `Right_Short => `Right_Long; `Left_Short => `Left_Long; // Division Rules: long cells divide to give raise to a long cell and a short cell `Right_Long => `Left_Long, `Right_Short; `Left_Long => `Left_Short, `Right_Long; } ;;
Let apply the evolution rules on the intitial state
S1 := grammar(S0) ;; S2 := grammar(S1) ;; S3 := grammar(S2) ;;
… which can be written shortly
S3 := grammar[iter = 3](S0) ;;
Let define a function to get a readable output
fun output(S) = ( foreach c in S do switch c case `Right_Short: stdout << "r" case `Left_Short: stdout << "l" case `Right_Long: stdout << "R" case `Left_Long: stdout << "L" endswitch; stdout << "\n"; S ) ;;
Let output data during the applications
grammar[iter = 5, prelude = output, interlude = output, postlude = output ](S0) ;;
Textual output of the program
R Lr lRR LLrLr lRlRRlRR LLrLLrLrLLrLr lRlRRlRlRRlRRlRlRRlRR LLrLLrLrLLrLLrLrLLrLrLLrLLrLrLLrLr lRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRR LLrLLrLrLLrLLrLrLLrLrLLrLLrLrLLrLLrLrLLrLrLLrLLrLrLLrLrLLrLLrLrLLrLLrLrLLrLrLLrLLrLrLLrLr lRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRRlRlRRlRlRRlRRlRlRRlRR
The whole code follows
- anabaena_growth_model.mgs
type cell = `Left_Long | `Right_Long | `Left_Short | `Right_Short ;; type anabaena = [cell]seq ;; S0 := seq:(`Right_Long) ;; anabaena(S0) ;; trans grammar = { `Right_Short => `Right_Long; `Left_Short => `Left_Long; `Right_Long => `Left_Long, `Right_Short; `Left_Long => `Left_Short, `Right_Long; } ;; fun output(S) = ( foreach c in S do switch c case `Right_Short: stdout << "r" case `Left_Short: stdout << "l" case `Right_Long: stdout << "R" case `Left_Long: stdout << "L" endswitch; stdout << "\n"; S ) ;; grammar[iter = 5, prelude = output, interlude = output, postlude = output ](S0) ;;
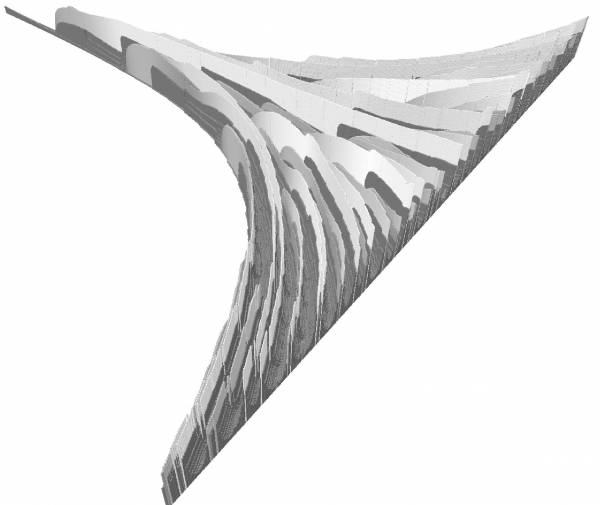
Heterocyst Differentiation Model
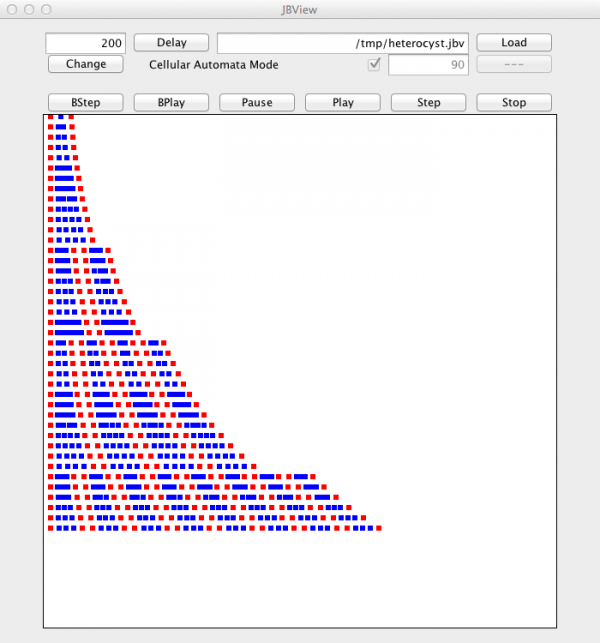
When starved for nitrogen, specialized cells called heterocysts differentiate from the photosynthetic vegetative cells at regular intervals along each filament. Heterocysts are anaerobic factories for nitrogen fixation; in them, the nitrogenase enzyme complex is synthesized and the components of the oxygen-evolving photosystem are turned off. Plant signals exert both positive and negative regulatory control on heterocyst differentiation.
Wilcox et al. have proposed a activator-inhibitor model of heterocysts differentiation where the (high) concentration of the activator triggers the heterocysts differentiation. The production of the activator is an autocatalytic reaction and also catalyzes the production of the inhibitor. The inhibitor is an antagonist substance that repress the activity of the activator when is concentration is high enough. The diffusion of the inhibitor to the neighboring cells prevents neighbors to become also heterocysts and explains why heterocysts appear in a regular spaced pattern in the filament.
The inhibitor have been identified more recently. The inhibition of the differentiation is a protein PatS which is produced by the heterocysts and which diffuses in the neighborbor vegetative cells. The model specified as a parametric L-system is presented in (Prusinkiewicz, 2012)1). The straightforward translation of this model in MGS follows.
MGS Implementation
- anabaena_heterocyst_differentiation.mgs
ofile := "heterocyst.jbv" ;; K := random(2.0) ;; v := 0.5 ;; R := 1.1 ;; T := 0.1 ;; Smax := 0.8 ;; dt := 0.001 ;; record W = { J: float } ;; record C = { a:`V|`H, c:float, s:float, p:`Right|`Left } ;; record H = C + { a = `H } ;; type anabaena = [W|C]seq ;; S0 := ({ a = `H, c = 1.0, s = 1.0, p = `Left }, { J = 0.0 }, { a = `V, c = 1.0, s = 1.0, p = `Right }, { J = 0.0 }, { a = `H, c = 1.0, s = 1.0, p = `Left }) ;; trans rules = { wall:W => { J = K * (left(wall).c - right(wall).c) } ; cell:H => cell; cell:C => let cell' = cell + { c = cell.c + dt * ((left(cell).J - right(cell).J) - v * cell.c), s = cell.s * (R ** dt) } in if cell'.c < T then cell' + { a = `H, c = 1.0, s = 1.0 } elif cell'.s > Smax then if cell'.p == `Right then (cell' + { s = 3.0 * cell'.s / 4.0, p = `Left }, { J = 0.0 }, cell' + { s = 1.0 * cell'.s / 4.0, p = `Right }) else (cell' + { s = 1.0 * cell'.s / 4.0, p = `Left }, { J = 0.0 }, cell' + { s = 3.0 * cell'.s / 4.0, p = `Right }) fi else cell' + { a = `V } fi } ;; fun output(S) = ( if iteration % 1000 == 0 then fold_indexed((fun p, c, acc -> if C(c) then ofile << " { " << (2.0 * acc + c.s) << " " << (iteration / 500); if c.a == `H then ofile << " 1 0 0 } " else ofile << " 0 0 1 } " fi; acc + c.s else acc fi), 0.0, S) fi; S );; ofile << " { ";; rules[iter = 40000, prelude = output, interlude = output, postlude = output ](S0) ;; ofile << " }\n" ;;